| Gene Symbol (Gene ID) | Ank3 (ENSMUSG00000069601) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cross Reference |
|
||||||||||||
| Definition |
protein_id:ENSMUSP00000138285
translation: MALPHSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLN RSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLV SFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRL VEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNG MDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASF PEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPS GEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLA DCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQE NFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQ EPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEP SMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLK KWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDG WQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVI PEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDA PKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
||||||||||||
| Chromosome | 10 | ||||||||||||
| Length Position(start - end) |
89456 69926176 - 70015633 |
||||||||||||
| Orient to Chromosome | plus | ||||||||||||
| Link | Location :
(10:69926176-70015633) / |
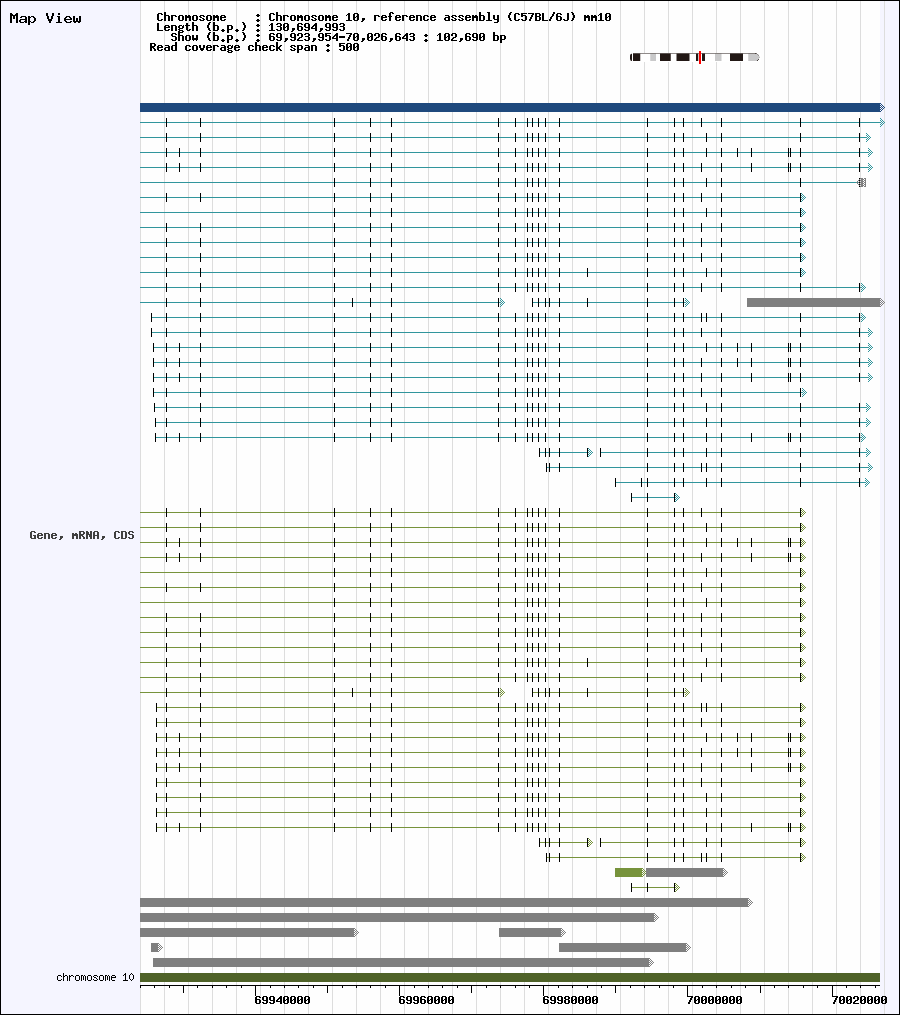
| Click to browse alignment view. |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | gene |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
locus_tag : Ank3 note : ankyrin 3, epithelial [Source:MGI Symbol;Acc:MGI:88026] |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182884 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182155 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000183169 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000183148 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182439 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000092434 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000047061 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000092432 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000092431 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000054167 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182992 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182972 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182683 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182692 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000092433 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182269 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000183261 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000183074 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182029 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000181974 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182795 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182437 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000186247 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000183082 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182207 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000183023 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000182665 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | mRNA |
| link |
VEGA
Ensembl MGI |
| xref | |
| definition |
note : transcript_id=ENSMUST00000185582 |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
CCDS:CCDS23909.1
Uniprot/SWISSPROT:G5E8K5 RefSeq_peptide:NP_666117 RefSeq_peptide:NP_733789.1 RefSeq_mRNA:NM_146005.3 RefSeq_mRNA:NM_170688.2 RefSeq_peptide_predicted:XP_011241617.1 Vega_transcript:Ank3-001 Vega_transcript:OTTMUST00000113771 UCSC:uc007fmx.1 Fantom:6720463O04 Fantom:9530086C08 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 EMBL:AK035681 EMBL:AK162007 EMBL:BC021657 EMBL:CH466553 EMBL:L40631 EMBL:L40632 EMBL:U89274 EMBL:U89275 GO:GO:0000281 GO:GO:0000281 GO:GO:0005200 GO:GO:0005200 GO:GO:0005515 GO:GO:0005764 GO:GO:0005856 GO:GO:0005923 GO:GO:0005923 GO:GO:0007009 GO:GO:0007009 GO:GO:0007165 GO:GO:0007409 GO:GO:0007411 GO:GO:0009925 GO:GO:0009925 GO:GO:0010960 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016323 GO:GO:0016328 GO:GO:0016328 GO:GO:0019228 GO:GO:0030424 GO:GO:0033268 GO:GO:0033270 GO:GO:0042383 GO:GO:0042383 GO:GO:0043001 GO:GO:0043001 GO:GO:0043194 GO:GO:0044325 GO:GO:0045184 GO:GO:0045184 GO:GO:0045202 GO:GO:0045211 GO:GO:0045760 GO:GO:0050808 GO:GO:0071286 GO:GO:0071709 GO:GO:0071709 GO:GO:0072659 GO:GO:0072659 GO:GO:0072660 GO:GO:0072660 GO:GO:0072661 GO:GO:0072661 GO:GO:1902260 MGI_trans_name:Ank3-001 Vega_translation:207718 Vega_translation:OTTMUSP00000063572 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005737 goslim_goa:GO:0005764 goslim_goa:GO:0005773 goslim_goa:GO:0005856 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0042592 goslim_goa:GO:0043226 goslim_goa:GO:0048856 goslim_goa:GO:0050877 goslim_goa:GO:0051301 goslim_goa:GO:0061024 protein_id:AAB01602.1 protein_id:AAB01603.1 protein_id:AAB01607.1 protein_id:AAB58380.1 protein_id:AAB58381.1 protein_id:AAH21657.1 protein_id:BAC29151.1 protein_id:BAE36677.1 protein_id:EDL31985.1 protein_id:EDL31989.1 protein_id:EDL31990.1 OTTP:OTTMUSP00000063572 OTTT:OTTMUST00000113771 UniParc:UPI0000D8AFEF |
| definition |
note : transcript_id=ENSMUST00000182884 protein_id : ENSMUSP00000138326 translation : MSEEPKEKPAKPAHRKRKGKKSDANASYLRAARAGHLEKALDYIKNGVDVNICNQNGLNALHLASKEGHVEVVSELLQREANVDAATKKGNTALHIASLAGQAEVVKVLVTNGANVNAQSQNGFTPLYMAAQENHLEVVRFLLDNGASQSLATEDGFTPLAVALQQGHDQVVSLLLENDTKGKVRLPALHIAARKDDTKAAALLLQNDTNADVESKSGFTPLHIAAHYGNINVATLLLNRAAAVDFTARNDITPLHVASKRGNANMVKLLLDRGAKIDAKTRDGLTPLHCGARSGHEQVVEMLLDRSAPILSKTKNGLSPLHMATQGDHLNCVQLLLQHNVPVDDVTNDYLTALHVAAHCGHYKVAKVLLDKKASPNAKALNGFTPLHIACKKNRIRVMELLLKHGASIQAVTESGLTPIHVAAFMGHVNIVSQLMHHGASPNTTNVRGETALHMAARSGQAEVVRYLVQDGAQVEAKAKDDQTPLHISARLGKADIVQQLLQQGASPNAATTSGYTPLHLAAREGHEDVAAFLLDHGASLSITTKKGFTPLHVAAKYGKLEVASLLLQKSASPDAAGKSGLTPLHVAAHYDNQKVALLLLDQGASPHAAAKNGYTPLHIAAKKNQMDIATSLLEYGADANAVTRQGIASVHLAAQEGHVDMVSLLLSRNANVNLSNKSGLTPLHLAAQEDRVNVAEVLVNQGAHVDAQTKMGYTPLHVGCHYGNIKIVNFLLQHSAKVNAKTKNGYTALHQAAQQGHTHIINVLLQNNASPNELTVNGNTALAIARRLGYISVVDTLKVVTEEIMTTTTITEKHKMNVPETMNEVLDMSDDEVRKASAPEKLSDGEYISDGEEGDKCTWFKIPKVQEVLVKSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSECAQCWASVPGIPNDGRQAEPLRPQTRKVGMSSEQQEKGKSGPDEEVTEDKVKSLFEDIQLEEVEAEEMTEDQGQAMLNRVQRAELAMSSLAGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
CCDS:CCDS59545.1
Uniprot/SWISSPROT:G5E8K5 RefSeq_peptide:NP_733924 RefSeq_mRNA:NM_170728.2 RefSeq_peptide_predicted:XP_006513201.1 Vega_transcript:Ank3-011 Vega_transcript:OTTMUST00000113793 UCSC:uc007fna.1 Fantom:6720463O04 Fantom:9530086C08 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 EMBL:AK035681 EMBL:AK162007 EMBL:BC021657 EMBL:CH466553 EMBL:L40631 EMBL:L40632 EMBL:U89274 EMBL:U89275 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005764 GO:GO:0005856 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007409 GO:GO:0007411 GO:GO:0009925 GO:GO:0010960 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0019228 GO:GO:0030424 GO:GO:0033268 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0044325 GO:GO:0045184 GO:GO:0045202 GO:GO:0045211 GO:GO:0045760 GO:GO:0050808 GO:GO:0071286 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 GO:GO:1902260 MGI_trans_name:Ank3-011 Vega_translation:207719 Vega_translation:OTTMUSP00000063578 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005737 goslim_goa:GO:0005764 goslim_goa:GO:0005773 goslim_goa:GO:0005856 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0042592 goslim_goa:GO:0043226 goslim_goa:GO:0048856 goslim_goa:GO:0050877 goslim_goa:GO:0051301 goslim_goa:GO:0061024 protein_id:AAB01602.1 protein_id:AAB01603.1 protein_id:AAB01607.1 protein_id:AAB58380.1 protein_id:AAB58381.1 protein_id:AAH21657.1 protein_id:BAC29151.1 protein_id:BAE36677.1 protein_id:EDL31985.1 protein_id:EDL31989.1 protein_id:EDL31990.1 OTTP:OTTMUSP00000063578 OTTT:OTTMUST00000113793 UniParc:UPI0000E59247 |
| definition |
note : transcript_id=ENSMUST00000182155 protein_id : ENSMUSP00000138347 translation : MSEEPKEKPAKPAHRKRKGKKSDANASYLRAARAGHLEKALDYIKNGVDVNICNQNGLNALHLASKEGHVEVVSELLQREANVDAATKKGNTALHIASLAGQAEVVKVLVTNGANVNAQSQNGFTPLYMAAQENHLEVVRFLLDNGASQSLATEDGFTPLAVALQQGHDQVVSLLLENDTKGKVRLPALHIAARKDDTKAAALLLQNDTNADVESKSGFTPLHIAAHYGNINVATLLLNRAAAVDFTARNDITPLHVASKRGNANMVKLLLDRGAKIDAKTRDGLTPLHCGARSGHEQVVEMLLDRSAPILSKTKNGLSPLHMATQGDHLNCVQLLLQHNVPVDDVTNDYLTALHVAAHCGHYKVAKVLLDKKASPNAKALNGFTPLHIACKKNRIRVMELLLKHGASIQAVTESGLTPIHVAAFMGHVNIVSQLMHHGASPNTTNVRGETALHMAARSGQAEVVRYLVQDGAQVEAKAKDDQTPLHISARLGKADIVQQLLQQGASPNAATTSGYTPLHLAAREGHEDVAAFLLDHGASLSITTKKGFTPLHVAAKYGKLEVASLLLQKSASPDAAGKSGLTPLHVAAHYDNQKVALLLLDQGASPHAAAKNGYTPLHIAAKKNQMDIATSLLEYGADANAVTRQGIASVHLAAQEGHVDMVSLLLSRNANVNLSNKSGLTPLHLAAQEDRVNVAEVLVNQGAHVDAQTKMGYTPLHVGCHYGNIKIVNFLLQHSAKVNAKTKNGYTALHQAAQQGHTHIINVLLQNNASPNELTVNGNTALAIARRLGYISVVDTLKVVTEEIMTTTTITEKHKMNVPETMNEVLDMSDDEGEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-014
Vega_transcript:OTTMUST00000113794 Uniprot/SPTREMBL:S4R1S2 UCSC:uc007fna.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-014 Vega_translation:207720 Vega_translation:OTTMUSP00000063579 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063579 OTTT:OTTMUST00000113794 UniParc:UPI0003335370 |
| definition |
note : transcript_id=ENSMUST00000183169 protein_id : ENSMUSP00000138348 translation : MSEEPKEKPAKPAHRKRKGKKSDANASYLRAARAGHLEKALDYIKNGVDVNICNQNGLNALHLASKEGHVEVVSELLQREANVDAATKKGNTALHIASLAGQAEVVKVLVTNGANVNAQSQNGFTPLYMAAQENHLEVVRFLLDNGASQSLATEDGFTPLAVALQQGHDQVVSLLLENDTKGKVRLPALHIAARKDDTKAAALLLQNDTNADVESKSGFTPLHIAAHYGNINVATLLLNRAAAVDFTARNDITPLHVASKRGNANMVKLLLDRGAKIDAKTRDGLTPLHCGARSGHEQVVEMLLDRSAPILSKTKNGLSPLHMATQGDHLNCVQLLLQHNVPVDDVTNDYLTALHVAAHCGHYKVAKVLLDKKASPNAKALNGFTPLHIACKKNRIRVMELLLKHGASIQAVTESGLTPIHVAAFMGHVNIVSQLMHHGASPNTTNVRGETALHMAARSGQAEVVRYLVQDGAQVEAKAKDDQTPLHISARLGKADIVQQLLQQGASPNAATTSGYTPLHLAAREGHEDVAAFLLDHGASLSITTKKGFTPLHVAAKYGKLEVASLLLQKSASPDAAGKSGLTPLHVAAHYDNQKVALLLLDQGASPHAAAKNGYTPLHIAAKKNQMDIATSLLEYGADANAVTRQGIASVHLAAQEGHVDMVSLLLSRNANVNLSNKSGLTPLHLAAQEDRVNVAEVLVNQGAHVDAQTKMGYTPLHVGCHYGNIKIVNFLLQHSAKVNAKTKNGYTALHQAAQQGHTHIINVLLQNNASPNELTVNGNTALAIARRLGYISVVDTLKVVTEEIMTTTTITEKHKMNVPETMNEVLDMSDDEGEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASPKISSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQVKSPGEAFTRMTACCYKDLRNSESDSSSEEEQRITTRVIRRRVIIKGEEAKNIPGESVTEEQFTDEEGNLITRKITRKVIRRIGPQERKQDDVQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-015
Vega_transcript:OTTMUST00000113795 Uniprot/SPTREMBL:S4R2S8 UCSC:uc007fnb.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-015 Vega_translation:207721 Vega_translation:OTTMUSP00000063580 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063580 OTTT:OTTMUST00000113795 UniParc:UPI0003335342 |
| definition |
note : transcript_id=ENSMUST00000183148 protein_id : ENSMUSP00000138770 translation : MSEEPKEKPAKPAHRKRKGKKSDANASYLRAARAGHLEKALDYIKNGVDVNICNQNGLNALHLASKEGHVEVVSELLQREANVDAATKKGNTALHIASLAGQAEVVKVLVTNGANVNAQSQNGFTPLYMAAQENHLEVVRFLLDNGASQSLATEDGFTPLAVALQQGHDQVVSLLLENDTKGKVRLPALHIAARKDDTKAAALLLQNDTNADVESKSGFTPLHIAAHYGNINVATLLLNRAAAVDFTARNDITPLHVASKRGNANMVKLLLDRGAKIDAKTRDGLTPLHCGARSGHEQVVEMLLDRSAPILSKTKNGLSPLHMATQGDHLNCVQLLLQHNVPVDDVTNDYLTALHVAAHCGHYKVAKVLLDKKASPNAKALNGFTPLHIACKKNRIRVMELLLKHGASIQAVTESGLTPIHVAAFMGHVNIVSQLMHHGASPNTTNVRGETALHMAARSGQAEVVRYLVQDGAQVEAKAKDDQTPLHISARLGKADIVQQLLQQGASPNAATTSGYTPLHLAAREGHEDVAAFLLDHGASLSITTKKGFTPLHVAAKYGKLEVASLLLQKSASPDAAGKSGLTPLHVAAHYDNQKVALLLLDQGASPHAAAKNGYTPLHIAAKKNQMDIATSLLEYGADANAVTRQGIASVHLAAQEGHVDMVSLLLSRNANVNLSNKSGLTPLHLAAQEDRVNVAEVLVNQGAHVDAQTKMGYTPLHVGCHYGNIKIVNFLLQHSAKVNAKTKNGYTALHQAAQQGHTHIINVLLQNNASPNELTVNGNTALAIARRLGYISVVDTLKVVTEEIMTTTTITEKHKMNVPETMNEVLDMSDDEGEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASPKISSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSECAQCWASVPGIPNDGRQAEPLRPQTRKVGMSSEQQEKGKSGPDEEVTEDKVKSLFEDIQLEEVEAEEMTEDQGQAMLNRVQRAELAMSSLAGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKDLRNSESDSSSEEEQRITTRVIRRRVIIKGEEAKNIPGESVTEEQFTDEEGNLITRKITRKVIRRIGPQERKQDDVQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
CCDS:CCDS23911.1
RefSeq_peptide:NP_733926 RefSeq_mRNA:NM_170730.2 Vega_transcript:Ank3-027 Vega_transcript:OTTMUST00000113842 Uniprot/SPTREMBL:G3X971 UCSC:uc007fnc.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 EMBL:CH466553 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-027 Vega_translation:207723 Vega_translation:OTTMUSP00000063595 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 protein_id:EDL31991.1 OTTP:OTTMUSP00000063595 OTTT:OTTMUST00000113842 UniParc:UPI0000434756 |
| definition |
note : transcript_id=ENSMUST00000182439 protein_id : ENSMUSP00000138356 translation : MSEEPKEKPAKPAHRKRKGKKHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
CCDS:CCDS23909.1
Uniprot/SWISSPROT:G5E8K5 RefSeq_peptide_predicted:XP_011241617.1 UCSC:uc007fmx.1 Fantom:6720463O04 Fantom:9530086C08 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 EMBL:AK035681 EMBL:AK162007 EMBL:BC021657 EMBL:CH466553 EMBL:L40631 EMBL:L40632 EMBL:U89274 EMBL:U89275 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005764 GO:GO:0005856 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007409 GO:GO:0007411 GO:GO:0009925 GO:GO:0010960 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0019228 GO:GO:0030424 GO:GO:0033268 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0044325 GO:GO:0045184 GO:GO:0045202 GO:GO:0045211 GO:GO:0045760 GO:GO:0050808 GO:GO:0071286 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 GO:GO:1902260 MGI_trans_name:Ank3-205 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005737 goslim_goa:GO:0005764 goslim_goa:GO:0005773 goslim_goa:GO:0005856 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0042592 goslim_goa:GO:0043226 goslim_goa:GO:0048856 goslim_goa:GO:0050877 goslim_goa:GO:0051301 goslim_goa:GO:0061024 protein_id:AAB01602.1 protein_id:AAB01603.1 protein_id:AAB01607.1 protein_id:AAB58380.1 protein_id:AAB58381.1 protein_id:AAH21657.1 protein_id:BAC29151.1 protein_id:BAE36677.1 protein_id:EDL31985.1 protein_id:EDL31989.1 protein_id:EDL31990.1 UniParc:UPI0000D8AFEF |
| definition |
note : transcript_id=ENSMUST00000092434 protein_id : ENSMUSP00000090090 translation : MSEEPKEKPAKPAHRKRKGKKSDANASYLRAARAGHLEKALDYIKNGVDVNICNQNGLNALHLASKEGHVEVVSELLQREANVDAATKKGNTALHIASLAGQAEVVKVLVTNGANVNAQSQNGFTPLYMAAQENHLEVVRFLLDNGASQSLATEDGFTPLAVALQQGHDQVVSLLLENDTKGKVRLPALHIAARKDDTKAAALLLQNDTNADVESKSGFTPLHIAAHYGNINVATLLLNRAAAVDFTARNDITPLHVASKRGNANMVKLLLDRGAKIDAKTRDGLTPLHCGARSGHEQVVEMLLDRSAPILSKTKNGLSPLHMATQGDHLNCVQLLLQHNVPVDDVTNDYLTALHVAAHCGHYKVAKVLLDKKASPNAKALNGFTPLHIACKKNRIRVMELLLKHGASIQAVTESGLTPIHVAAFMGHVNIVSQLMHHGASPNTTNVRGETALHMAARSGQAEVVRYLVQDGAQVEAKAKDDQTPLHISARLGKADIVQQLLQQGASPNAATTSGYTPLHLAAREGHEDVAAFLLDHGASLSITTKKGFTPLHVAAKYGKLEVASLLLQKSASPDAAGKSGLTPLHVAAHYDNQKVALLLLDQGASPHAAAKNGYTPLHIAAKKNQMDIATSLLEYGADANAVTRQGIASVHLAAQEGHVDMVSLLLSRNANVNLSNKSGLTPLHLAAQEDRVNVAEVLVNQGAHVDAQTKMGYTPLHVGCHYGNIKIVNFLLQHSAKVNAKTKNGYTALHQAAQQGHTHIINVLLQNNASPNELTVNGNTALAIARRLGYISVVDTLKVVTEEIMTTTTITEKHKMNVPETMNEVLDMSDDEVRKASAPEKLSDGEYISDGEEGDKCTWFKIPKVQEVLVKSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSECAQCWASVPGIPNDGRQAEPLRPQTRKVGMSSEQQEKGKSGPDEEVTEDKVKSLFEDIQLEEVEAEEMTEDQGQAMLNRVQRAELAMSSLAGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
CCDS:CCDS23911.1
Uniprot/SPTREMBL:G3X971 UCSC:uc007fnc.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 EMBL:CH466553 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-201 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 protein_id:EDL31991.1 UniParc:UPI0000434756 |
| definition |
note : transcript_id=ENSMUST00000047061 protein_id : ENSMUSP00000045834 translation : MSEEPKEKPAKPAHRKRKGKKHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
CCDS:CCDS48590.1
RefSeq_peptide:NP_733790 RefSeq_mRNA:NM_170689.2 RefSeq_peptide_predicted:XP_006513197.1 RefSeq_peptide_predicted:XP_006513198.1 Uniprot/SPTREMBL:G5E8K3 UCSC:uc007fmy.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 EMBL:CH466553 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-204 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 protein_id:EDL31984.1 UniParc:UPI0000E59248 |
| definition |
note : transcript_id=ENSMUST00000092432 protein_id : ENSMUSP00000090088 translation : MSEEPKEKPAKPAHRKRKGKKSDANASYLRAARAGHLEKALDYIKNGVDVNICNQNGLNALHLASKEGHVEVVSELLQREANVDAATKKGNTALHIASLAGQAEVVKVLVTNGANVNAQSQNGFTPLYMAAQENHLEVVRFLLDNGASQSLATEDGFTPLAVALQQGHDQVVSLLLENDTKGKVRLPALHIAARKDDTKAAALLLQNDTNADVESKSGFTPLHIAAHYGNINVATLLLNRAAAVDFTARNDITPLHVASKRGNANMVKLLLDRGAKIDAKTRDGLTPLHCGARSGHEQVVEMLLDRSAPILSKTKNGLSPLHMATQGDHLNCVQLLLQHNVPVDDVTNDYLTALHVAAHCGHYKVAKVLLDKKASPNAKALNGFTPLHIACKKNRIRVMELLLKHGASIQAVTESGLTPIHVAAFMGHVNIVSQLMHHGASPNTTNVRGETALHMAARSGQAEVVRYLVQDGAQVEAKAKDDQTPLHISARLGKADIVQQLLQQGASPNAATTSGYTPLHLAAREGHEDVAAFLLDHGASLSITTKKGFTPLHVAAKYGKLEVASLLLQKSASPDAAGKSGLTPLHVAAHYDNQKVALLLLDQGASPHAAAKNGYTPLHIAAKKNQMDIATSLLEYGADANAVTRQGIASVHLAAQEGHVDMVSLLLSRNANVNLSNKSGLTPLHLAAQEDRVNVAEVLVNQGAHVDAQTKMGYTPLHVGCHYGNIKIVNFLLQHSAKVNAKTKNGYTALHQAAQQGHTHIINVLLQNNASPNELTVNGNTALAIARRLGYISVVDTLKVVTEEIMTTTTITEKHKMNVPETMNEVLDMSDDEVRKASAPEKLSDGEYISDGEEGEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSECAQCWASVPGIPNDGRQAEPLRPQTRKVGMSSEQQEKGKSGPDEEVTEDKVKSLFEDIQLEEVEAEEMTEDQGQAMLNRVQRAELAMSSLAGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
CCDS:CCDS23910.1
RefSeq_peptide:NP_733791 RefSeq_mRNA:NM_170690.2 Uniprot/SPTREMBL:G5E8K2 UCSC:uc007fmz.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 EMBL:CH466553 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-203 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 protein_id:EDL31994.1 UniParc:UPI0000D8AFF0 |
| definition |
note : transcript_id=ENSMUST00000092431 protein_id : ENSMUSP00000090087 translation : MSEEPKEKPAKPAHRKRKGKKSDANASYLRAARAGHLEKALDYIKNGVDVNICNQNGLNALHLASKEGHVEVVSELLQREANVDAATKKGNTALHIASLAGQAEVVKVLVTNGANVNAQSQNGFTPLYMAAQENHLEVVRFLLDNGASQSLATEDGFTPLAVALQQGHDQVVSLLLENDTKGKVRLPALHIAARKDDTKAAALLLQNDTNADVESKSGFTPLHIAAHYGNINVATLLLNRAAAVDFTARNDITPLHVASKRGNANMVKLLLDRGAKIDAKTRDGLTPLHCGARSGHEQVVEMLLDRSAPILSKTKNGLSPLHMATQGDHLNCVQLLLQHNVPVDDVTNDYLTALHVAAHCGHYKVAKVLLDKKASPNAKALNGFTPLHIACKKNRIRVMELLLKHGASIQAVTESGLTPIHVAAFMGHVNIVSQLMHHGASPNTTNVRGETALHMAARSGQAEVVRYLVQDGAQVEAKAKDDQTPLHISARLGKADIVQQLLQQGASPNAATTSGYTPLHLAAREGHEDVAAFLLDHGASLSITTKKGFTPLHVAAKYGKLEVASLLLQKSASPDAAGKSGLTPLHVAAHYDNQKVALLLLDQGASPHAAAKNGYTPLHIAAKKNQMDIATSLLEYGADANAVTRQGIASVHLAAQEGHVDMVSLLLSRNANVNLSNKSGLTPLHLAAQEDRVNVAEVLVNQGAHVDAQTKMGYTPLHVGCHYGNIKIVNFLLQHSAKVNAKTKNGYTALHQAAQQGHTHIINVLLQNNASPNELTVNGNTALAIARRLGYISVVDTLKVVTEEIMTTTTITEKHKMNVPETMNEVLDMSDDEGDKCTWFKIPKVQEVLVKSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSECAQCWASVPGIPNDGRQAEPLRPQTRKVGMSSEQQEKGKSGPDEEVTEDKVKSLFEDIQLEEVEAEEMTEDQGQAMLNRVQRAELAMSSLAGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
CCDS:CCDS35931.1
Uniprot/SWISSPROT:G5E8K5 RefSeq_peptide:NP_733925 RefSeq_mRNA:NM_170729.2 RefSeq_peptide_predicted:XP_006513199.1 RefSeq_peptide_predicted:XP_006513200.1 UCSC:uc007fnb.1 Fantom:6720463O04 Fantom:9530086C08 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 EMBL:AK035681 EMBL:AK162007 EMBL:BC021657 EMBL:CH466553 EMBL:L40631 EMBL:L40632 EMBL:U89274 EMBL:U89275 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005764 GO:GO:0005856 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007409 GO:GO:0007411 GO:GO:0009925 GO:GO:0010960 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0019228 GO:GO:0030424 GO:GO:0033268 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0044325 GO:GO:0045184 GO:GO:0045202 GO:GO:0045211 GO:GO:0045760 GO:GO:0050808 GO:GO:0071286 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 GO:GO:1902260 MGI_trans_name:Ank3-202 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005737 goslim_goa:GO:0005764 goslim_goa:GO:0005773 goslim_goa:GO:0005856 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0042592 goslim_goa:GO:0043226 goslim_goa:GO:0048856 goslim_goa:GO:0050877 goslim_goa:GO:0051301 goslim_goa:GO:0061024 protein_id:AAB01602.1 protein_id:AAB01603.1 protein_id:AAB01607.1 protein_id:AAB58380.1 protein_id:AAB58381.1 protein_id:AAH21657.1 protein_id:BAC29151.1 protein_id:BAE36677.1 protein_id:EDL31985.1 protein_id:EDL31989.1 protein_id:EDL31990.1 UniParc:UPI0000E59249 |
| definition |
note : transcript_id=ENSMUST00000054167 protein_id : ENSMUSP00000061698 translation : MSEEPKEKPAKPAHRKRKGKKSDANASYLRAARAGHLEKALDYIKNGVDVNICNQNGLNALHLASKEGHVEVVSELLQREANVDAATKKGNTALHIASLAGQAEVVKVLVTNGANVNAQSQNGFTPLYMAAQENHLEVVRFLLDNGASQSLATEDGFTPLAVALQQGHDQVVSLLLENDTKGKVRLPALHIAARKDDTKAAALLLQNDTNADVESKSGFTPLHIAAHYGNINVATLLLNRAAAVDFTARNDITPLHVASKRGNANMVKLLLDRGAKIDAKTRDGLTPLHCGARSGHEQVVEMLLDRSAPILSKTKNGLSPLHMATQGDHLNCVQLLLQHNVPVDDVTNDYLTALHVAAHCGHYKVAKVLLDKKASPNAKALNGFTPLHIACKKNRIRVMELLLKHGASIQAVTESGLTPIHVAAFMGHVNIVSQLMHHGASPNTTNVRGETALHMAARSGQAEVVRYLVQDGAQVEAKAKDDQTPLHISARLGKADIVQQLLQQGASPNAATTSGYTPLHLAAREGHEDVAAFLLDHGASLSITTKKGFTPLHVAAKYGKLEVASLLLQKSASPDAAGKSGLTPLHVAAHYDNQKVALLLLDQGASPHAAAKNGYTPLHIAAKKNQMDIATSLLEYGADANAVTRQGIASVHLAAQEGHVDMVSLLLSRNANVNLSNKSGLTPLHLAAQEDRVNVAEVLVNQGAHVDAQTKMGYTPLHVGCHYGNIKIVNFLLQHSAKVNAKTKNGYTALHQAAQQGHTHIINVLLQNNASPNELTVNGNTALAIARRLGYISVVDTLKVVTEEIMTTTTITEKHKMNVPETMNEVLDMSDDEGEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSECAQCWASVPGIPNDGRQAEPLRPQTRKVGMSSEQQEKGKSGPDEEVTEDKVKSLFEDIQLEEVEAEEMTEDQGQAMLNRVQRAELAMSSLAGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
RefSeq_peptide_predicted:XP_006513196.1
Vega_transcript:Ank3-005 Vega_transcript:OTTMUST00000113774 Uniprot/SPTREMBL:S4R2K9 UCSC:uc007fmy.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-005 Vega_translation:207724 Vega_translation:OTTMUSP00000063573 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063573 OTTT:OTTMUST00000113774 UniParc:UPI00033353E2 |
| definition |
note : transcript_id=ENSMUST00000182992 protein_id : ENSMUSP00000138686 translation : MAHAASQLKKNRDLEINAEEETEKKRKHRKRSRDRKKKSDANASYLRAARAGHLEKALDYIKNGVDVNICNQNGLNALHLASKEGHVEVVSELLQREANVDAATKKGNTALHIASLAGQAEVVKVLVTNGANVNAQSQNGFTPLYMAAQENHLEVVRFLLDNGASQSLATEDGFTPLAVALQQGHDQVVSLLLENDTKGKVRLPALHIAARKDDTKAAALLLQNDTNADVESKMVVNRATESGFTPLHIAAHYGNINVATLLLNRAAAVDFTARNDITPLHVASKRGNANMVKLLLDRGAKIDAKTRDGLTPLHCGARSGHEQVVEMLLDRSAPILSKTKNGLSPLHMATQGDHLNCVQLLLQHNVPVDDVTNDYLTALHVAAHCGHYKVAKVLLDKKASPNAKALNGFTPLHIACKKNRIRVMELLLKHGASIQAVTESGLTPIHVAAFMGHVNIVSQLMHHGASPNTTNVRGETALHMAARSGQAEVVRYLVQDGAQVEAKAKDDQTPLHISARLGKADIVQQLLQQGASPNAATTSGYTPLHLAAREGHEDVAAFLLDHGASLSITTKKGFTPLHVAAKYGKLEVASLLLQKSASPDAAGKSGLTPLHVAAHYDNQKVALLLLDQGASPHAAAKNGYTPLHIAAKKNQMDIATSLLEYGADANAVTRQGIASVHLAAQEGHVDMVSLLLSRNANVNLSNKSGLTPLHLAAQEDRVNVAEVLVNQGAHVDAQTKMGYTPLHVGCHYGNIKIVNFLLQHSAKVNAKTKNGYTALHQAAQQGHTHIINVLLQNNASPNELTVNGNTALAIARRLGYISVVDTLKVVTEEIMTTTTITEKHKMNVPETMNEVLDMSDDEVRKASAPEKLSDGEYISDGEEGEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMKTVERSSGTARSLPTTYSHKPFFSTRPYQSWTTAPITVPGPAKSGSLSSSPSNTPSASPLKSIWSVSTPSPIKSTLGASTTSSVKSISDVASPIRSFRTVSSPIKTVVSPSPYNPQVASGTLGRVPTITEATPIKGLAPNSTFSSRTSPVTTAGSLLERSSITMTPPASPKSNITMYSSSLPFKSIITSATPLISSPLKSVVSPTKSAADVISTAKATMASSLSSPLKQMSGHAEVALVNGSVSPLKYPSSSALINGCKATATLQDKISTATNAVSSVVSAASDTVEKALSTTTAMPFSPLRSYVSAAPSAFQSLRTPSASALYTSLGSSIAATTSSVTSSIITVPVYSVVNVLPEPALKKLPDSNSFTKSAAALLSPIKTLTTETRPQPHFNRTSSPVKSSLFLASSALKPSVPSSLSSSQEILKDVAEMKEDLMRMTAILQTDVPEEKPFQTDLPREGRIDDEEPFKIVEKVKEDLVKVSEILKKDVCVESKGPPKSPKSDKGHSPEDDWTEFSSEEIREARQAAASHAPSLPERVHGKANLTRVIDYLTNDIGSSSLTNLKYKFEEAKKDGEERQKRILKPAMALQEHKLKMPPASMRPSTSEKELCKMADSFFGADAILESPDDFSQHDQDKSPLSDSGFETRSEKTPSAPQSAESTGPKPLFHEVPIPPVITETRTEVVHVIRSYEPSSGEIPQSQPEDPVSPKPSPTFMELEPKPTTSSIKEKVKAFQMKASSEEEDHSRVLSKGMRVKEETHITTTTRMVYHSPPGGECASERIEETMSVHDIMKAFQSGRDPSKELAGLFEHKSAMSPDVAKSAAETSAQHAEKDSQMKPKLERIIEVHIEKGPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-016
Vega_transcript:OTTMUST00000113800 Uniprot/SPTREMBL:S4R236 UCSC:uc007fnk.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-016 Vega_translation:207727 Vega_translation:OTTMUSP00000063583 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063583 OTTT:OTTMUST00000113800 UniParc:UPI0003335384 |
| definition |
note : transcript_id=ENSMUST00000182972 protein_id : ENSMUSP00000138481 translation : KTKNGYTALHQAAQQGHTHIINVLLQNNASPNELTVNGNTALAIARRLGYISVVDTLKVVTEEIMTTTTITEKHKMNVPETMNEVLDMSDDEGEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSECAQCWASVPGIPNDGRQAEPLRPQTRKVGMSSEQQEKGKSGPDEEVTEDKVKSLFEDIQLEEVEAEEMTEDQGQAMLNRVQRAELAMSSLAGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-034
Vega_transcript:OTTMUST00000113857 Uniprot/SPTREMBL:S4R1U4 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-034 Vega_translation:207728 Vega_translation:OTTMUSP00000063605 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063605 OTTT:OTTMUST00000113857 UniParc:UPI0003335377 |
| definition |
note : transcript_id=ENSMUST00000182683 protein_id : ENSMUSP00000138375 translation : EEIMTTTTITEKHKMNVPETMNEVLDMSDDEGEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGCSSPLPQYDSRFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQA |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-030
Vega_transcript:OTTMUST00000113843 Uniprot/SPTREMBL:S4R2F5 UCSC:uc007fnk.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-030 Vega_translation:207729 Vega_translation:OTTMUSP00000063596 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063596 OTTT:OTTMUST00000113843 UniParc:UPI00033353B0 |
| definition |
note : transcript_id=ENSMUST00000182692 protein_id : ENSMUSP00000138623 translation : MALPHSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
CCDS:CCDS23912.1
Uniprot/SWISSPROT:G5E8K5 RefSeq_peptide:NP_733788 RefSeq_mRNA:NM_170687.3 Vega_transcript:Ank3-018 Vega_transcript:OTTMUST00000113802 UCSC:uc007fnk.1 Fantom:6720463O04 Fantom:9530086C08 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 EMBL:AK035681 EMBL:AK162007 EMBL:BC021657 EMBL:CH466553 EMBL:L40631 EMBL:L40632 EMBL:U89274 EMBL:U89275 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005764 GO:GO:0005856 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007409 GO:GO:0007411 GO:GO:0009925 GO:GO:0010960 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0019228 GO:GO:0030424 GO:GO:0033268 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0044325 GO:GO:0045184 GO:GO:0045202 GO:GO:0045211 GO:GO:0045760 GO:GO:0050808 GO:GO:0071286 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 GO:GO:1902260 MGI_trans_name:Ank3-018 Vega_translation:207730 Vega_translation:OTTMUSP00000063584 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005737 goslim_goa:GO:0005764 goslim_goa:GO:0005773 goslim_goa:GO:0005856 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0042592 goslim_goa:GO:0043226 goslim_goa:GO:0048856 goslim_goa:GO:0050877 goslim_goa:GO:0051301 goslim_goa:GO:0061024 protein_id:AAB01602.1 protein_id:AAB01603.1 protein_id:AAB01607.1 protein_id:AAB58380.1 protein_id:AAB58381.1 protein_id:AAH21657.1 protein_id:BAC29151.1 protein_id:BAE36677.1 protein_id:EDL31985.1 protein_id:EDL31989.1 protein_id:EDL31990.1 OTTP:OTTMUSP00000063584 OTTT:OTTMUST00000113802 UniParc:UPI0000692459 |
| definition |
note : transcript_id=ENSMUST00000092433 protein_id : ENSMUSP00000090089 translation : MALPHSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSECAQCWASVPGIPNDGRQAEPLRPQTRKVGMSSEQQEKGKSGPDEEVTEDKVKSLFEDIQLEEVEAEEMTEDQGQAMLNRVQRAELAMSSLAGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-019
Vega_transcript:OTTMUST00000113803 Uniprot/SPTREMBL:S4R187 UCSC:uc007fnj.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-019 Vega_translation:207731 Vega_translation:OTTMUSP00000063585 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063585 OTTT:OTTMUST00000113803 UniParc:UPI0003335212 |
| definition |
note : transcript_id=ENSMUST00000182269 protein_id : ENSMUSP00000138123 translation : MALPHSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASPKISSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKVKSPGEAFTRMTACCYKDLRNSESDSSSEEEQRITTRVIRRRVIIKGEEAKNIPGESVTEEQFTDEEGNLITRKITRKVIRRIGPQERKQDDVQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-020
Vega_transcript:OTTMUST00000113844 Uniprot/SPTREMBL:S4R165 UCSC:uc007fnk.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-020 Vega_translation:207732 Vega_translation:OTTMUSP00000063597 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063597 OTTT:OTTMUST00000113844 UniParc:UPI00033353A3 |
| definition |
note : transcript_id=ENSMUST00000183261 protein_id : ENSMUSP00000138095 translation : MALPHSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASPKISSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSECAQCWASVPGIPNDGRQAEPLRPQTRKVGMSSEQQEKGKSGPDEEVTEDKVKSLFEDIQLEEVEAEEMTEDQGQAMLNRVQRAELAMSSLAGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKVKSPGEAFTRMTACCYKDLRNSESDSSSEEEQRITTRVIRRRVIIKGEEAKNIPGESVTEEQFTDEEGNLITRKITRKVIRRIGPQERKQDDVQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-021
Vega_transcript:OTTMUST00000113845 Uniprot/SPTREMBL:S4R2J6 UCSC:uc007fnk.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-021 Vega_translation:207733 Vega_translation:OTTMUSP00000063598 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063598 OTTT:OTTMUST00000113845 UniParc:UPI0003335380 |
| definition |
note : transcript_id=ENSMUST00000183074 protein_id : ENSMUSP00000138671 translation : MALPHSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASPKISSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSECAQCWASVPGIPNDGRQAEPLRPQTRKVGMSSEQQEKGKSGPDEEVTEDKVKSLFEDIQLEEVEAEEMTEDQGQAMLNRVQRAELAMSSLAGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKDLRNSESDSSSEEEQRITTRVIRRRVIIKGEEAKNIPGESVTEEQFTDEEGNLITRKITRKVIRRIGPQERKQDDVQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Uniprot/SWISSPROT:G5E8K5
RefSeq_peptide_predicted:XP_006513202.1 Vega_transcript:Ank3-022 Vega_transcript:OTTMUST00000113846 UCSC:uc007fni.1 Fantom:6720463O04 Fantom:9530086C08 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 EMBL:AK035681 EMBL:AK162007 EMBL:BC021657 EMBL:CH466553 EMBL:L40631 EMBL:L40632 EMBL:U89274 EMBL:U89275 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005764 GO:GO:0005856 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007409 GO:GO:0007411 GO:GO:0009925 GO:GO:0010960 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0019228 GO:GO:0030424 GO:GO:0033268 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0044325 GO:GO:0045184 GO:GO:0045202 GO:GO:0045211 GO:GO:0045760 GO:GO:0050808 GO:GO:0071286 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 GO:GO:1902260 MGI_trans_name:Ank3-022 Vega_translation:207734 Vega_translation:OTTMUSP00000063599 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005737 goslim_goa:GO:0005764 goslim_goa:GO:0005773 goslim_goa:GO:0005856 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0042592 goslim_goa:GO:0043226 goslim_goa:GO:0048856 goslim_goa:GO:0050877 goslim_goa:GO:0051301 goslim_goa:GO:0061024 protein_id:AAB01602.1 protein_id:AAB01603.1 protein_id:AAB01607.1 protein_id:AAB58380.1 protein_id:AAB58381.1 protein_id:AAH21657.1 protein_id:BAC29151.1 protein_id:BAE36677.1 protein_id:EDL31985.1 protein_id:EDL31989.1 protein_id:EDL31990.1 OTTP:OTTMUSP00000063599 OTTT:OTTMUST00000113846 UniParc:UPI000287A2E5 |
| definition |
note : transcript_id=ENSMUST00000182029 protein_id : ENSMUSP00000138337 translation : MALPHSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSECAQCWASVPGIPNDGRQAEPLRPQTRKVGMSSEQQEKGKSGPDEEVTEDKVKSLFEDIQLEEVEAEEMTEDQGQAMLNRVQRAELAMSSLAGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
CCDS:CCDS59546.1
Uniprot/SWISSPROT:G5E8K5 RefSeq_peptide:NP_033800 RefSeq_mRNA:NM_009670.4 Vega_transcript:Ank3-024 Vega_transcript:OTTMUST00000113848 UCSC:uc007fnj.1 Fantom:6720463O04 Fantom:9530086C08 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 EMBL:AK035681 EMBL:AK162007 EMBL:BC021657 EMBL:CH466553 EMBL:L40631 EMBL:L40632 EMBL:U89274 EMBL:U89275 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005764 GO:GO:0005856 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007409 GO:GO:0007411 GO:GO:0009925 GO:GO:0010960 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0019228 GO:GO:0030424 GO:GO:0033268 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0044325 GO:GO:0045184 GO:GO:0045202 GO:GO:0045211 GO:GO:0045760 GO:GO:0050808 GO:GO:0071286 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 GO:GO:1902260 MGI_trans_name:Ank3-024 Vega_translation:207735 Vega_translation:OTTMUSP00000063600 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005737 goslim_goa:GO:0005764 goslim_goa:GO:0005773 goslim_goa:GO:0005856 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0042592 goslim_goa:GO:0043226 goslim_goa:GO:0048856 goslim_goa:GO:0050877 goslim_goa:GO:0051301 goslim_goa:GO:0061024 protein_id:AAB01602.1 protein_id:AAB01603.1 protein_id:AAB01607.1 protein_id:AAB58380.1 protein_id:AAB58381.1 protein_id:AAH21657.1 protein_id:BAC29151.1 protein_id:BAE36677.1 protein_id:EDL31985.1 protein_id:EDL31989.1 protein_id:EDL31990.1 OTTP:OTTMUSP00000063600 OTTT:OTTMUST00000113848 UniParc:UPI0000027F8D |
| definition |
note : transcript_id=ENSMUST00000181974 protein_id : ENSMUSP00000138285 translation : MALPHSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASLRSFSSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-025
Vega_transcript:OTTMUST00000113849 Uniprot/SPTREMBL:S4R1X7 UCSC:uc007fnj.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-025 Vega_translation:207736 Vega_translation:OTTMUSP00000063601 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063601 OTTT:OTTMUST00000113849 UniParc:UPI000333529E |
| definition |
note : transcript_id=ENSMUST00000182795 protein_id : ENSMUSP00000138413 translation : MALPHSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-026
Vega_transcript:OTTMUST00000113850 Uniprot/SPTREMBL:S4R2C1 UCSC:uc007fnj.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-026 Vega_translation:207737 Vega_translation:OTTMUSP00000063602 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063602 OTTT:OTTMUST00000113850 UniParc:UPI00033353BC |
| definition |
note : transcript_id=ENSMUST00000182437 protein_id : ENSMUSP00000138586 translation : MALPHSEDAITGDTDKYLGPQDLKELGDDSLPAEGYVGFSLGARSASPKISSDRSYTLNRSSYARDSMMIEELLVPSKEQHLTFTREFDSDSLRHYSWAADTLDNVNLVSSPVHSGFLVSFMVDARGGSMRGSRHHGMRIIIPPRKCTAPTRITCRLVKRHKLANPPPMVEGEGLASRLVEMGPAGAQFLGPVIVEIPHFGSMRGKERELIVLRSENGETWKEHQFDSKNEDLAELLNGMDEELDSPEELGTKRICRIITKDFPQYFAVVSRIKQESNQIGPEGGILSSTTVPLVQASFPEGALTKRIRVGLQAQPVPEETVKKILGNKATFSPIVTVEPRRRKFHKPITMTIPVPPPSGEGVSNGYKGDATPNLRLLCSITGGTSPAQWEDITGTTPLTFIKDCVSFTTNVSARFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKAEKADRRQSFASLALRPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKDLRNSESDSSSEEEQRITTRVIRRRVIIKGEEAKNIPGESVTEEQFTDEEGNLITRKITRKVIRRIGPQERKQDDVQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-038
Vega_transcript:OTTMUST00000124840 Uniprot/SPTREMBL:A0A087WNU5 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-038 Vega_translation:207738 Vega_translation:OTTMUSP00000067834 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000067834 OTTT:OTTMUST00000124840 UniParc:UPI00046210E7 |
| definition |
codon_start : 2 note : transcript_id=ENSMUST00000186247 protein_id : ENSMUSP00000139496 translation : XFWLADCHQVLETVGLASQLYRELICVPYMAKFVVFAKTNDPVESSLRCFCMTDDRVDKTLEQQENFEEVARSKDIEVLEGKPIYVDCYGNLAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKETESDQDDEAEKADRRQSFASLALRKRYSYLTEPSMIERSSGTARSLPTTYSHKPFFSTRPYQSWTTAPITVPGPAKSGSLSSSPSNTPSASPLKSIWSVSTPSPIKSTLGASTTSSVKSISDVASPIRSFRTVSSPIKTVVSPSPYNPQVASGTLGRVPTITEATPIKGLAPNSTFSSRTSPVTTAGSLLERSSITMTPPASPKSNITMYSSSLPFKSIITSATPLISSPLKSVVSPTKSAADVISTAKATMASSLSSPLKQMSGHAEVALVNGSVSPLKYPSSSALINGCKATATLQDKISTATNAVSSVVSAASDTVEKALSTTTAMPFSPLRSYVSAAPSAFQSLRTPSASALYTSLGSSIAATTSSVTSSIITVPVYSVVNVLPEPALKKLPDSNSFTKSAAALLSPIKTLTTETRPQPHFNRTSSPVKSSLFLASSALKPSVPSSLSSSQEILKDVAEMKEDLMRMTAILQTDVPEEKPFQTDLPREGRIDDEEPFKIVEKVKEDLVKVSEILKKDVCVESKGPPKSPKSDKGHSPEDDWTEFSSEEIREARQAAASHAPSLPERVHGKANLTRVIDYLTNDIGSSSLTNLKYKFEEAKKDGEERQKRILKPAMALQEHKLKMPPASMRPSTSEKELCKMADSFFGADAILESPDDFSQHDQDKSPLSDSGFETRSEKTPSAPQSAESTGPKPLFHEVPIPPVITETRTEVVHVIRSYEPSSGEIPQSQPEDPVSPKPSPTFMELEPKPTTSSIKEKVKAFQMKASSEEEDHSRVLSKGMRVKEETHITTTTRMVYHSPPGGECASERIEETMSVHDIMKAFQSGRDPSKELAGLFEHKSAMSPDVAKSAAETSAQHAEKDSQMKPKLERIIEVHIEKGNQAEPTEVIIRETKKHPEKEVSVYQKDLSRGNINLRDFLPEKHDAFPCPEEQGQQEEEELAAEESLPSYLESSRVNTPGSQEEDSRPSSAQLLSDDSYKTLKLLSQHSVEYHDDELSDLRGESYRFAEKMLLSEKLDVSHSDTEESVTDHAGPPSSELQGSDKRSREKVATAPPKEILSKIYKDVSENGLGRVSKDEHFEKLTVLHYSGNVSGPKHAMWMHLSEDRLDRGREKLMYEDRVDRTVKEAEEKLTEVSQFFRDKTEKLNDELQSPEKKPRPKNGKDYSSQSSTSSSPEKVLTELLASNDEWVKARQRGPDGQSVPQAEDRKAPSRSNSPENRVPTQQSEDDQPPEEAKRTVVAQSRGQEGPQSGFQLKQSKLSSIRLKFEQGARAKSKDPPHEEKHLDGPSRIPVKKTQETKLPTHPGFAREKQQKAVDPLEERVPVQNDVTVFKADHAQSNEIVTSKSGSGNGKSHRTEMLSKAMPDFFPEQQVEDSACPITSDLETKGPWDRKVFRTWESSGANNTKAQKEQLSHVLVHDIRENHAGRPDDSENGDPKSGFMYVTEREHKMLTNGSLSEIKEMSVKSPSKKVLYREYIVKDGDPPSSALNHPPRRSESSLASHIPIRVTDERRMLSSNIPDGFCEQSTFPKQELSPRVSRPSMSEGVVESQHFNSVDDEKVTYSEISKVSKHQSYLALDETETSPTKSPDSLEFSPGKDSPSSDVFDHGSVDGLEKTEGGKEIKTLPVYVSFVQVGKQYEKELQPGGVKKIISQECKTVQEARGTFYTARQQKQPPSPQGSPEDDTLEQVSFLDSSGKSPLTPETPSSEEVSYEFTSKTPDSLIAFIPGQPSPIPEVSEESEEEEPKSAPLRQVTVEKETDRDVSKDSIQRPKCNRVAYIEFPPPPPLDADQMESDKKHQYLPEREVDMMEVSLQDESDKYQLAEPVIRVQPPSPVPPGAEASDSSDDESLYQPVPVKKYTFKLKEVDEGQKDTAKSKTAATKASNQKEADGNGREGESGLDSPQNETAQNGNNDQSVTECSIATTAEFSHDTDATEIDSLDGYDLQDEDDGLTESDSKLPSQTIDTKKDVWTEGILKPADRSFSQSKLEVIEEEEGKVGLDEEKPSPSKSPSSDRTPEKADPKSGAQFFTLEGRHPDRSVLPDTYFSYKVDEEFATPFKTVATKGLDFDPWPNNRGDNEVFDGKSREDDTKPFGLAVDDRSPATTPDTTPARTPTDESTPTSEPNPFPFHEGKMFEMTRSGAIDMSKRDFVEERLQFFQIGEHTPEGKSGAQGEGDMVTDTPQPQSGDTSVDTNLERDVVAPSVDPNPSIPSNGECQEGTACSGSLEKSAAATNTSKVDPKSRTPIKMGISASTMTMKKEGSGEVTDKTEAVVTSCQGLENEIVKEISSAPSSQVGIRPQEKHDFQKDNFNNNNNLDASTMQTDNSTSHIVLTGRAASTCTTEEANPVKGSGKSPGTQGHSSRESRKEPIGLRRKSKLPIKATAPKDVFPPNHKADSKTGKPRQVGQYEKHKALPTSSCLDAKSRIPVKNTHRENLVSVRKACATQKRGQPERGKAKQPPSKLPVKVRSTRVTVTTTNTSTTTTTTTTTTTTTTVKVTESQLKEVCKHPIEYFKGISGETLKLVDRLTEEDKKMQSELSDEEESTSRNTSLSETSRGGQPSVTTKSARNKKTEAPPLKSKRGKAGSRRTGPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVD |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-035
Vega_transcript:OTTMUST00000113859 Uniprot/SPTREMBL:S4R162 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-035 Vega_translation:207739 Vega_translation:OTTMUSP00000063606 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063606 OTTT:OTTMUST00000113859 UniParc:UPI000333528A |
| definition |
codon_start : 2 note : transcript_id=ENSMUST00000183082 protein_id : ENSMUSP00000138092 translation : XAPLTKGGQQLVFNFYSFKENRLPFSIKIRDTSQEPCGRLSFLKEPKTTKGLPQTAVCNLNITLPAHKKETESDQDDEAEKADRRQSFASLALRKRYSYLTEPSMKTVERSSGT |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-031
Vega_transcript:OTTMUST00000113860 Uniprot/SPTREMBL:S4R278 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-031 Vega_translation:207740 Vega_translation:OTTMUSP00000063607 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063607 OTTT:OTTMUST00000113860 UniParc:UPI00033353CE |
| definition |
note : transcript_id=ENSMUST00000182207 protein_id : ENSMUSP00000138531 translation : PKTTKGLPQTAVCNLNITLPAHKKETESDQDDEAEKADRRQSFASLALRKRYSYLTEPSMSPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGHPSFQVELETPMGLYCTPPNPFQQDDHFSDISSIESPFRTPSRLSDGLVPSQGNIEHPTGGPPVVTAEDTSLEDSKMDDSVTVTDPADPLDVDESQLKDLCQSWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKQGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-006
Vega_transcript:OTTMUST00000113775 Uniprot/SPTREMBL:S4R208 UCSC:uc007fnm.1 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-006 Vega_translation:207741 Vega_translation:OTTMUSP00000063574 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000063574 OTTT:OTTMUST00000113775 UniParc:UPI00033353EF |
| definition |
note : transcript_id=ENSMUST00000183023 protein_id : ENSMUSP00000138450 translation : EHKLKMPPASMRPSTSEKELCKMADSFFGADAILESPDDFSQHDQDKSPLSDSGFETRSEKTPSAPQSAESTGPKPLFHEVPIPPVITETRTEVVHVIRSYEPSSGEIPQSQPEDPVSPKPSPTFMELEPKPTTSSIKEKVKAFQMKASSEEEDHSRVLSKGMRVKEETHITTTTRMVYHSPPGGECASERIEETMSVHDIMKAFQSGRDPSKELAGLFEHKSAMSPDVAKSAAETSAQHAEKDSQMKPKLERIIEVHIEKGPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLISQSFMLLKKWVTRDGKNATTDALTSVLTKINRIDIVTLLEGPIFDYGNISGTRSFADENNVFHDPVDGWQNETPSGSLESPAQARRLTGGLLDRLDDSSDQARDSITSYLTGEPGKIEANGNHTAEVIPEAKAKPYFPESQNDIGKQSIKENLKPKTHGCGRTEEPVSPLTAYQKSLEETSKLVIEDAPKPCVPVGMKKMTRTTADGKARLNLQEEEGSTRSEPKGEGYKVKTKKEIRNVEKKTH |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-029
Vega_transcript:OTTMUST00000113852 Uniprot/SPTREMBL:A0A087WRU2 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-029 Vega_translation:207742 Vega_translation:OTTMUSP00000067835 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000067835 OTTT:OTTMUST00000113852 UniParc:UPI0004620ED0 |
| definition |
codon_start : 3 note : transcript_id=ENSMUST00000182665 protein_id : ENSMUSP00000140773 translation : XVDPLEERVPVQNDVTVFKADHAQSNEIVTSKSGSGNGKSHRTEMLSKAMPDFFPEQQVEDSACPITSDLETKGPWDRKVFRTWESSGANNTKAQKEQLSHVLVHDIRENHAGRPDDSENGDPKSGFMYVTEREHKMLTNGSLSEIKEMSVKSPSKKVLYREYIVKDGDPPSSALNHPPRRSESSLASHIPIRVTDERRMLSSNIPDGFCEQSTFPKQELSPRVSRPSMSEGVVESQHFNSVDDEKVTYSEISKVSKHQSYLALDETETSPTKSPDSLEFSPGKDSPSSDVFDHGSVDGLEKTEGGKEIKTLPVYVSFVQVGKQYEKELQPGGVKKIISQECKTVQEARGTFYTARQQKQPPSPQGSPEDDTLEQVSFLDSSGKSPLTPETPSSEEVSYEFTSKTPDSLIAFIPGQPSPIPEVSEESEEEEPKSAPLRQVTVEKETDRDVSKDSIQRPKCNRVAYIEFPPPPPLDADQMESDKKHQYLPEREVDMMEVSLQDESDKYQLAEPVIRVQPPSPVPPGAEASDSSDDESLYQPVPVKKYTFKLKEVDEGQKDTAKSKTAATKASNQKEADGNGREGESGLDSPQNETAQNGNNDQSVTECSIATTAEFSHDTDATEIDSLDGYDLQDEDDGLTESDSKLPSQTIDTKKDVWTEGILKPADRSFSQSKLEVIEEEEGKVGLDEEKPSPSKSPSSDRTPEKADPKSGAQFFTLEGRHPDRSVLPDTYFSYKVDEEFATPFKTVATKGLDFDPWPNNRGDNEVFDGKSREDDTKPFGLAVDDRSPATTPDTTPARTPTDESTPTSEPNPFPFHEGKMFEMTRSGAIDMSKRDFVEERLQFFQIVARP |
| name | Ank3 (ENSMUSG00000069601) (Detail) |
|---|---|
| type | CDS |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:Ank3-039
Vega_transcript:OTTMUST00000124841 Uniprot/SPTREMBL:A0A087WRP9 EMBL:AC100427 EMBL:AC129018 EMBL:AC132435 EMBL:AC156836 GO:GO:0000281 GO:GO:0005200 GO:GO:0005515 GO:GO:0005622 GO:GO:0005923 GO:GO:0007009 GO:GO:0007165 GO:GO:0007411 GO:GO:0009925 GO:GO:0014704 GO:GO:0016020 GO:GO:0016323 GO:GO:0016328 GO:GO:0030424 GO:GO:0033270 GO:GO:0042383 GO:GO:0043001 GO:GO:0043194 GO:GO:0045184 GO:GO:0045202 GO:GO:0050808 GO:GO:0071709 GO:GO:0072659 GO:GO:0072660 GO:GO:0072661 MGI_trans_name:Ank3-039 Vega_translation:207743 Vega_translation:OTTMUSP00000067836 goslim_goa:GO:0000902 goslim_goa:GO:0003674 goslim_goa:GO:0005198 goslim_goa:GO:0005575 goslim_goa:GO:0005622 goslim_goa:GO:0005623 goslim_goa:GO:0005886 goslim_goa:GO:0006605 goslim_goa:GO:0006810 goslim_goa:GO:0007009 goslim_goa:GO:0007049 goslim_goa:GO:0007165 goslim_goa:GO:0008150 goslim_goa:GO:0016192 goslim_goa:GO:0022607 goslim_goa:GO:0030154 goslim_goa:GO:0040011 goslim_goa:GO:0048856 goslim_goa:GO:0051301 goslim_goa:GO:0061024 OTTP:OTTMUSP00000067836 OTTT:OTTMUST00000124841 UniParc:UPI00046211DF |
| definition |
note : transcript_id=ENSMUST00000185582 protein_id : ENSMUSP00000140724 translation : GRHPDRSVLPDTYFSYKVDEEFATPFKTVATKGLDFDPWPNNRGDNEVFDGKSREDDTKPFGLAVDDRSPATTPDTTPARTPTDESTPTSEPNPFPFHEGKMFEMTRSGAIDMSKRDFVEERLQFFQIGPQSPCERTDIRMAIVADHLGLSWTELARELNFSVDEINQIRVENPNSLI |
| name | |
|---|---|
| type | STS |
| link | |
| xref |
UniSTS_NUM:185834
UniSTS:L40632 WI_MRC-RH:4906 |
| definition |
note : map_weight=1 standard_name : L40632 |
| name | |
|---|---|
| type | STS |
| link | |
| xref |
UniSTS_NUM:141079
UniSTS:Ank3 |
| definition |
note : map_weight=2 standard_name : Ank3 |
| name | |
|---|---|
| type | misc_feature |
| link | |
| xref | |
| definition |
note : contig AC156836.6 1..225328(-1) |
| name | |
|---|---|
| type | misc_feature |
| link | |
| xref | |
| definition |
note : contig AC132435.3 1..197120(-1) |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | misc_RNA |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:OTTMUST00000113772
Vega_transcript:OTTMUST00000113772 MGI_trans_name:Ank3-003 OTTT:OTTMUST00000113772 |
| definition |
note : retained_intron note : transcript_id=ENSMUST00000182474 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | misc_RNA |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:OTTMUST00000113858
Vega_transcript:OTTMUST00000113858 MGI_trans_name:Ank3-033 OTTT:OTTMUST00000113858 |
| definition |
note : retained_intron note : transcript_id=ENSMUST00000182239 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | misc_RNA |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:OTTMUST00000113801
Vega_transcript:OTTMUST00000113801 MGI_trans_name:Ank3-017 OTTT:OTTMUST00000113801 |
| definition |
note : retained_intron note : transcript_id=ENSMUST00000182367 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | misc_RNA |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:OTTMUST00000113847
Vega_transcript:OTTMUST00000113847 MGI_trans_name:Ank3-023 OTTT:OTTMUST00000113847 |
| definition |
note : retained_intron note : transcript_id=ENSMUST00000182687 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | misc_RNA |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:OTTMUST00000113851
Vega_transcript:OTTMUST00000113851 MGI_trans_name:Ank3-028 OTTT:OTTMUST00000113851 |
| definition |
note : retained_intron note : transcript_id=ENSMUST00000182762 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | misc_RNA |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:OTTMUST00000113861
Vega_transcript:OTTMUST00000113861 MGI_trans_name:Ank3-037 OTTT:OTTMUST00000113861 |
| definition |
note : retained_intron note : transcript_id=ENSMUST00000182606 |
| name | Ank3 (ENSMUSG00000069601) |
|---|---|
| type | misc_RNA |
| link |
VEGA
Ensembl MGI |
| xref |
Vega_transcript:OTTMUST00000113862
Vega_transcript:OTTMUST00000113862 MGI_trans_name:Ank3-036 OTTT:OTTMUST00000113862 |
| definition |
note : retained_intron note : transcript_id=ENSMUST00000183001 |


